The analysis of bone marrow smears (BMS) serves as a critical diagnostic tool in hematology, offering valuable insights into the cellular composition and distinctive morphology of bone marrow cells. It enables the accurate diagnosis, classification, and monitoring of various hematologic diseases. Currently, both clinical routine diagnostics and scientific research, especially for the implementation of digital technologies, rely heavily on the manual selection of regions of interest (ROI) suitable for cell-level evaluation. Current guidelines recommend the manual evaluation of up to 500 cells per sample making the process time-consuming, cost-ineffective, subjective, and prone to human error. Consequently, significant inter-observer variability and inconsistencies in diagnosis often arise.
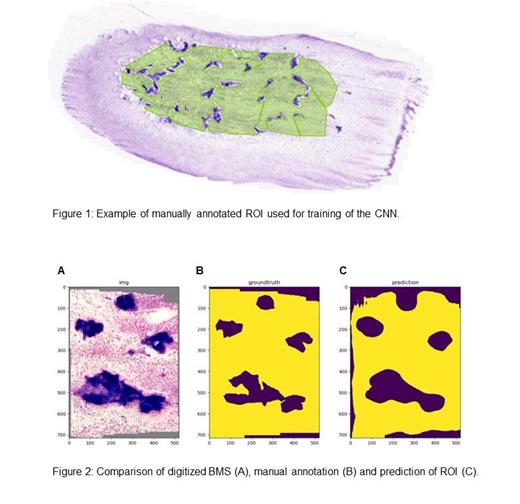
Automated analysis of BMS using whole slide images (WSI) offers a promising approach to alleviate financial and time-related burdens while promoting standardization and streamlining of routine diagnostics. We have developed a machine learning approach utilizing a fully convolutional neural network (CNN) specifically designed for semantic segmentation based on the algorithm by Long et al. (IEEE Computer Society, 2015). The model is capable of learning deep semantic representations in its classification backbone combined with shape features in a shallow layer to produce detailed segmentations. The classification backbone consists of an adapted residual neural network with 50 layers (Resnet50) by He et al. (arXiv:1512.03385, 2015). To train the model, we curated a diverse dataset consisting of 60 different WSI BMS, each manually and independently annotated by at least two experts providing a robust ground truth (figure 1). Data augmentation was performed via horizontal flipping, random resizing, and random cropping. A train-test split of 80:20 was implemented with 5-fold internal cross-validation. The model's accuracy was evaluated as mean intersection-over-union ratio yielding high congruency between ground truth and model prediction (figure 2). The final model uses downscaled images effectively reducing data storage size and data transfer time decreasing fully automated ROI prediction time to <300 ms for one WSI.
The model allows for in-depth examination at the cellular level, harnessing the potential of thousands of relevant cells present in a WSI, in contrast to the limited range of 200 to 500 cells typically examined in conventional clinical routine diagnostics. Thereby, the model solves an essential bottleneck in computer vision in microscopy minimizing human manual labor and standardizing BMS processing results.
Disclosures
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal